Incorporating an Unnatural Amino Acid To Improve NMR - JACS Spotlight
Incorporating an Unnatural Amino Acid To Improve NMR
Christen Brownlee
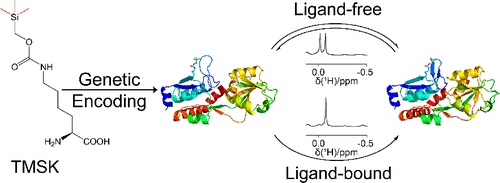
The enzyme pyrrolysyl-tRNA synthetase (PyIRS) and its cognate pyrrolysyl-tRNA have been used extensively to add unnatural amino acids to the genetic code of bacterial and eukaryotic cells. However, despite the potential of this system for studying proteins, protein yields of most pyIRS variants have been generally modest, limiting their application in biomolecular NMR.Gottfried Otting and co-workers show new potential for this system, using it for site-specific incorporation of a high-signal lysine derivative into proteins with high yield (DOI: 10.1021/jacs.0c11971). This variant, (((trimethylsilyl)methoxy)carbonyl)-l-lysine (TMSK), consists of a trimethylsilyl group at the end of a lysine side chain. With nine equivalent protons, TMSK is able to produce intense singlets in 1H NMR spectra near 0 ppm, where few other proton resonances occur. The researchers demonstrate this system’s ability to detect ligand binding, measure the rate of conformational change, and assess protein dimerization by paramagnetic relaxation enhancement. They also show the ability to incorporate both TMSK and O-tert-butyl-tyrosine, another unnatural amino acid, in the same protein in quantities sufficient for NMR spectroscopy. The close proximity of these two noncanonical amino acids was readily detected by nuclear Overhauser effects. The authors suggest that small and flexible probes like TMSK have excellent potential for site-specific protein studies.
